CC-Radiomics-Phantom-3 | CT Phantom Scans for Head, Chest, and Controlled Protocols on 100 Scanners
DOI: 10.7937/tcia.2019.j71i4fah | Data Citation Required | 204 Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Head, Chest, and Phantom | Phantom | 95 | RTSTRUCT, CT, Protocol, Other | Phantom | Image Analyses | Public, Complete | 2019/05/08 |
Summary
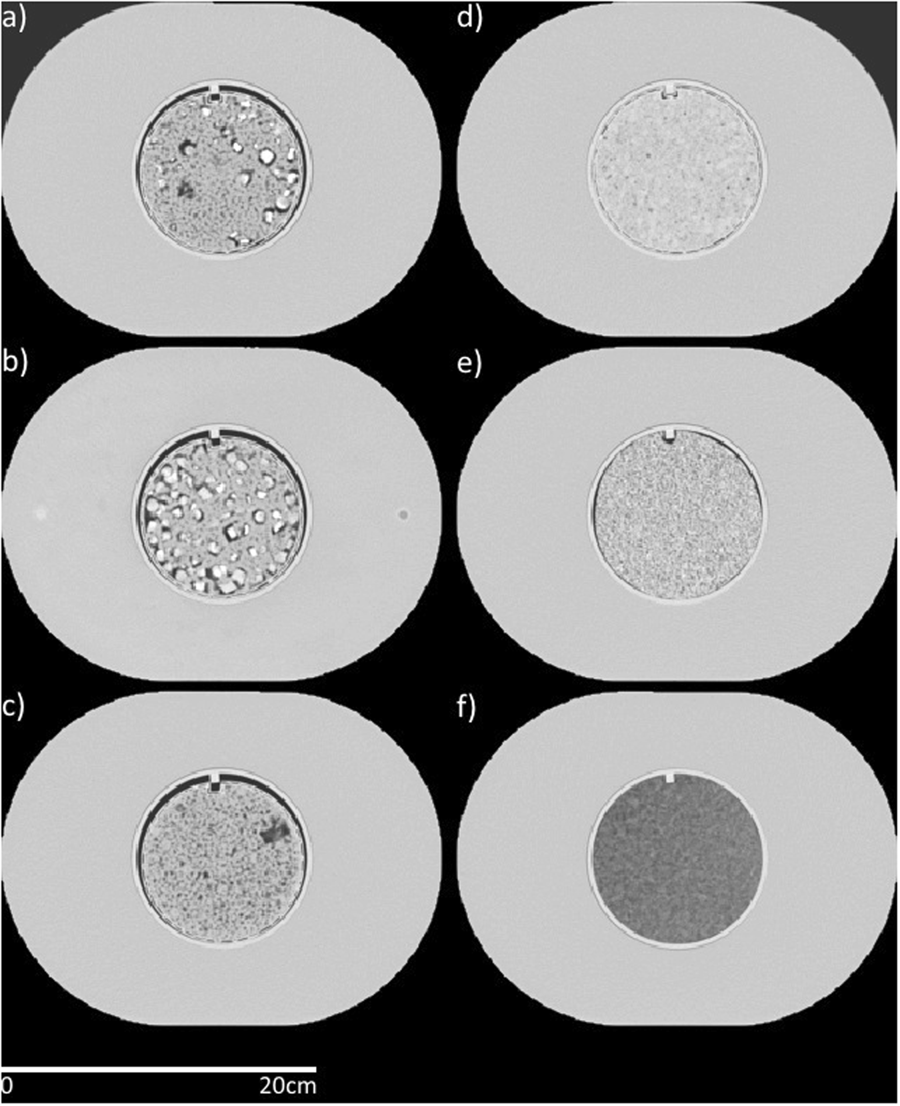
There currently is a dearth of phantom scans on large samples. This data collection contains one physical phantom, imaged across three protocols, on 100 scanners. This provides population data that can be used to quantify inter-scanner variability. This data can be used to determine how robust specific radiomics or other quantitative imaging signatures are. Computed tomography scans were acquired on 100 scanners at 35 clinics: 51 GE scanners, 20 Philips scanners, 11 Toshiba scanners, and 1 Philips and Neusoft Medical System scanner. The commonly used chest and head protocols of the local clinic were acquired without changing the protocol parameters. Additionally, a controlled protocol was acquired that was designed to minimize radiomics feature differences between manufacturers. The settings for the controlled protocol were: tube voltage, 120 kV(p); tube current, 200 mA∙s; helical scan type; spiral pitch factor, 1.0; 50-cm display field of view; and image thickness, 3 mm (except for GE scanners, which used an image thickness of 2.5 mm). The convolution kernel was standard for GE; C for Philips; B31f, B31s for Siemens; and FC08 for Toshiba. However, the kernel used for the Toshiba scans switched from FC18 (six scanners) to FC08 (five scanners) halfway through owing to a study by Mackin et al. (Medical Physics, 2018) that found the FC08 kernel to match the GE standard kernel best. K-means clustering showed that the scanners did not cluster by kernel type, thus all Toshiba scanners were included. There were 94 scanners that had a controlled protocol scan that could be used: 48 GE, 18 Philips, 17 Siemens, and 11 Toshiba scanners; 93 scanners that had a local chest protocol scan that could be used: 47 GE, 19 Philips, 17 Siemens, and 10 Toshiba scanners; and 88 scanners that had a local head protocol scan that could be used: 46 GE, 18 Philips, 14 Siemens, and 10 Toshiba scanners. The various reasons that scans could not be used were as follows: the field of view did not encompass all the cartridges, the scan extent did not cover the length of the phantom, and the scan was acquired with variable image thickness. The featured image shows Axial views from a computed tomography scan of the radiomics phantom used. The cartridges are (a) 50% acrylonitrile butadiene styrene (ABS), 25% acrylic beads, and 25% polyvinyl chloride (PVC) pieces (percentages are by weight), (b) 50% ABS and 50% PVC pieces, (c) 50% ABS and 50% acrylic beads, (d) hemp seeds in polyurethane, (e) shredded rubber, and (f) dense cork. Protocol
The high-density polystyrene buildup is seen outside the cartridges with dimensions of 28cm×21cm×22cm. The cartridges had a diameter of 10.8 cm. Window width: 1600, window level: -300.
Data Access
Version 1: Updated 2019/05/08
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images and Radiation Therapy Structures | RTSTRUCT, CT | DICOM | Download requires NBIA Data Retriever |
95 | 113 | 550 | 25,612 | CC BY 3.0 | View |
| Acquisition & reconstruction settings | Protocol, Other | XLSX | CC BY 3.0 | — |
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Ger R, Zhou S, Chi P-C, Lee H, Layman R, Jones K, Goff DL, Fuller CD, Howell RM, Heng L, Stafford RJ, Court LE, Mackin D. (2019). Data from CT Phantom Scans for Head, Chest, and Controlled Protocols on 100 Scanners (CC-Radiomics-Phantom-3) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/tcia.2019.j71i4fah |
Detailed Description
Note:
Acquisition and reconstruction settings within the DICOM header may be incorrect. The settings used for each scan are provided in the attached spreadsheet.)
Acknowledgements
This data set was provided to TCIA by Rachel Ger, Shouhao Zhou, Pai-Chun Chi, Hannah Lee, Rick Layman, Kyle Jones, David Goff, Carlos Cardenas, Clifton Fuller, Rebecca Howell, Heng Li, Jason Stafford, Laurence Court, Dennis Mackin.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Ger, Rachel B.; Zhou, Shouhao; Chi, Pai-Chun Melinda; Lee, Hannah J.; Layman, Rick R.; Jones, A. Kyle; Goff, David L.; Fuller, Clifton D.; Howell, Rebecca M.; Li, Heng; Stafford, R. Jason; Court, Laurence E.; Mackin, Dennis S. (2018) Comprehensive Investigation on Controlling for CT Imaging Variabilities in Radiomics Studies. Scientific Reports 8:13047. DOI: 10.1038/s41598-018-31509-z |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
