Lung-Fused-CT-Pathology | Fused Radiology-Pathology Lung Dataset
DOI: 10.7937/k9/tcia.2018.smt36lpn | Data Citation Required | 732 Views | 2 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Lung | Human | 6 | CT, SEG, Histopathology, Demographic, Measurement, Other | Lung Cancer | Image Analyses, Software/Source Code | Public, Complete | 2018/07/30 |
Summary
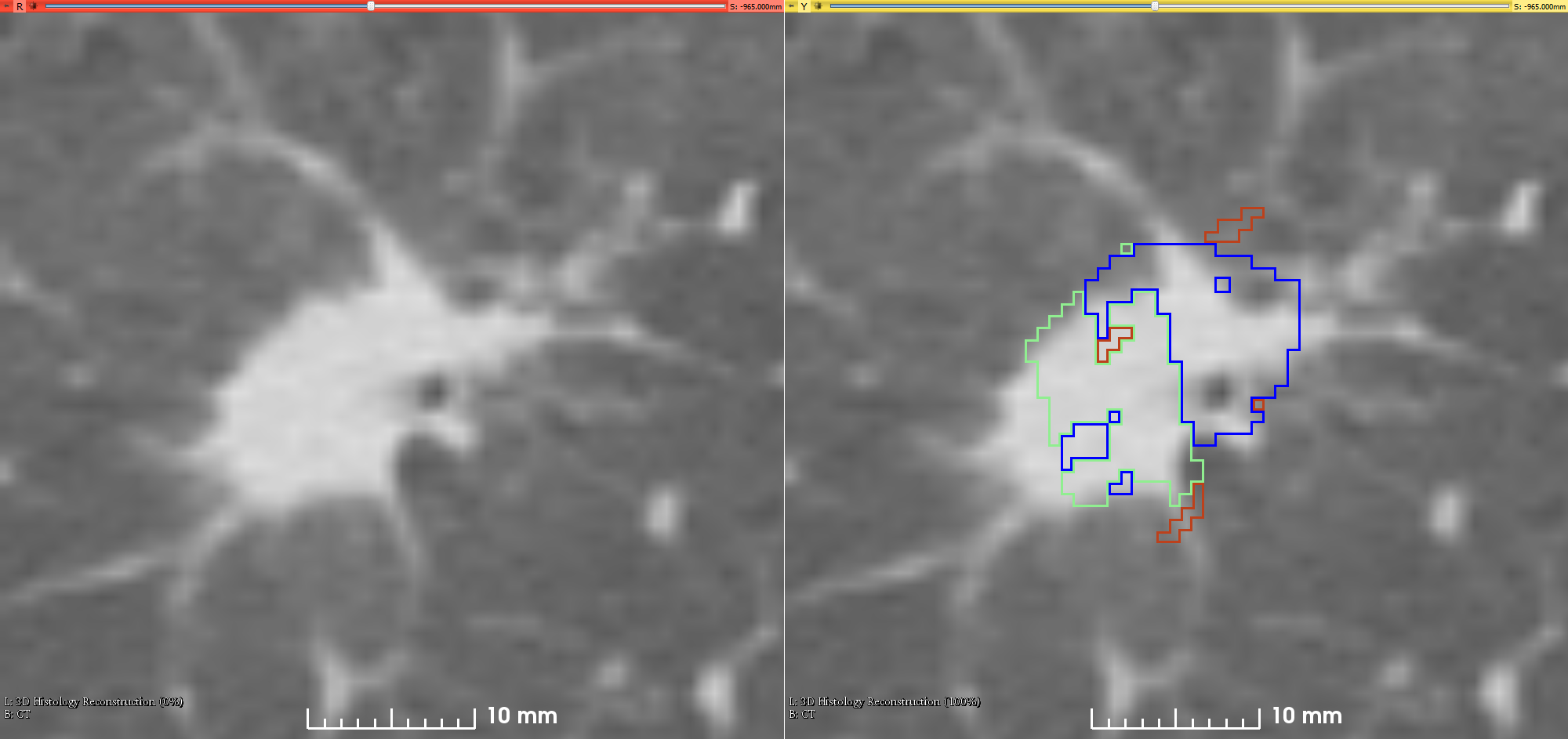
This is the first attempt of mapping the extent of Invasive Adenocarcinoma onto in vivo lung CT. The mappings constitute ground truth of disease and may be used to further investigate the imaging signatures of Invasive Adenocarcinoma in ground glass pulmonary nodules. Patient with small ground glass nodules with >2 histology slices per nodule were included. Patients with solid large nodules (>40mm), with <3 histology slices or with histology slices showing substantial artifacts were excluded from this study (see reference below for details). Data collection and analysis was provided by Case Western Reserve University. References
Data Access
Version 1: Updated 2018/07/30
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| CT Images and Histology compartments mapped on CT | CT, SEG | DICOM | Download requires NBIA Data Retriever |
6 | 36 | 52 | 11,210 | CC BY 3.0 | View |
| Annotated Whole Slide Pathology Images | Histopathology | TIFF | Download requires IBM-Aspera-Connect plugin |
6 | 6 | 25 | CC BY 3.0 | — | |
| Clinical data | Demographic, Measurement, Other | XLSX | CC BY 3.0 | — |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
- Source code is publicly available on Github at https://github.com/mirabelarusu/RadPathFusionLung
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Madabhushi, A., & Rusu, M. (2018). Fused Radiology-Pathology Lung (Lung-Fused-CT-Pathology) (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/k9/tcia.2018.smt36lpn |
Detailed Description
Supporting Documentation
The data set is fully described in the following publications:
Rusu et al. Co-registration of pre-operative CT with ex vivo surgically excised ground glass nodules to define spatial extent of invasive adenocarcinoma on in vivo imaging: a proof-of-concept study. European Radiology (2018); PMCID:PMC5630490 DOI:10.1007/s00330-017-4813-0
Histology Data Description
There is one folder for each patient, with the same folder name as the TCIA ID. Each folder with TCIA ID name contains 2 folders:
- “images”
- Contains scanned histology images (in tiff format) pertinent to the TCIA ID Patient, e.g. LungFCP-01-0001_b1.tiff, LungFCP-01-0001_b2.tiff, etc
- “annotations”
- Contains the pathologist’s annotations, stored as tiff file with the same image size as the scanned histology files, with 0 where there is not label, and 255 where a region was annotated. It is possible to have multiple annotations for each file, e.g. for file LungFCP-01-0001\images\LungFCP-01-0001_b1.tiff the following regions are available:
- LungFCP-01-0001_b1_annotation_00_R000G255B000.tiff
- LungFCP-01-0001_b1_annotation_00_R255G000B000.tiff
- Contains the pathologist’s annotations, stored as tiff file with the same image size as the scanned histology files, with 0 where there is not label, and 255 where a region was annotated. It is possible to have multiple annotations for each file, e.g. for file LungFCP-01-0001\images\LungFCP-01-0001_b1.tiff the following regions are available:
A part of the filenames (LungFCP-01-0001_b1_annotation_00_RxxxGyyyBzzz) indicate the type of annotation:
- Adenocarcinoma in Situ : R000G000B255, R001G000B255, R002G000B255
- Invasive Adenocarcinoma: R000G255B000, R001G255B000, R002G255B000, R003G255B000, R004G255B000, R005G255B000, R006G255B000
- Invasive Adenocarcinoma + Adenocarcinoma in Situ: R255G000B000
If multiple files are available for the same type of annotation, e.g. Invasive Adenocarcinoma, it indicates that the pathologists has annotated multiple regions
Content of “FinalPublishedResults” folder
The folder “FinalPublishedResults” contains one folder for each patient. Within each patient folder, there are 3 subfolders: “CT”, ”Histology”, & ”Results_XX” where XX is a number. When expanded each patient folder appears as depicted below.
- LungFCP-01-0001
- CT
- CT.mha
CT_crop_blood_label.mha
CT_label_Meta_crop.mha
- CT.mha
- histology
- imgs
- masks
- blood
- invasive
- lesion
- sample
- (airways)
- Results_XX
- blood_label.mha
blood_label_up.mha
blood_label_up_bin.mha
invasion_label.mha
invasion_label_up.mha
invasion_label_up_bin.mha
labels_up_bin.mha
lesion.mha
lesion_label.mha
lesion_label_up.mha
lesion_label_up_bin.mha
reconstruction_grayscale.mha
sample_label.mha
- blood_label.mha
- CT
- LungFCP-01-0002
- LungFCP-01-0003
- LungFCP-01-0004
- LungFCP-01-0005
- LungFCP-01-0006
The folder “CT” contains the 3D CT volume saved as one mha file, the segmentation of the lesion obtained by majority voting for the three radiologist segmentations, and the segmentation of the blood vessels.
The folder “histology” has a similar content as the raw histology data, but a lower resolution, with some additional processing, e.g. applied gross rotation and flipping to correct for artifacts related to the mounting on the glass slide, and with additional annotations beside the lesion and in situ disease, e.g. blood vessels.
The folder Results_XX contain the outcome of the registration of the histopathology images and the CT.
All scripts used for generating these results are available at https://github.com/mirabelarusu/RadPathFusionLung
Please refer to the following publication for the methodological details:
Rusu M., Rajiah P., Gilkeson R., Yang M., Donatelli C., Thawani R., Jacono F.J., Linden P., Madabushi A. (2017) Co-registration of pre-operative CT with ex vivo surgically excised ground glass nodules to define spatial extent of invasive adenocarcinoma on in vivo imaging: a proof-of-concept study. European Radiology 27:10, 4209:4217. DOI: https://doi.org/10.1007/s00330-017-4813-0
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Rusu, M., Rajiah, P., Gilkeson, R., Yang, M., Donatelli, C., Thawani, R., Jacono, F. J., Linden, P., & Madabhushi, A. (2018) Co-registration of pre-operative CT with ex vivo surgically excised ground glass nodules to define spatial extent of invasive adenocarcinoma on in vivo imaging: a proof-of-concept study. European Radiology (Vol. 27, Issue 10, pp. 4209–4217). PMCID:PMC5630490 DOI: https://doi.org/10.1007/s00330-017-4813-0 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
