GammaKnife-Hippocampal | Gamma Knife MR/CT/RTSTRUCT Sets With Hippocampal Contours
DOI: 10.7937/Q967-X166 | Data Citation Required | 426 Views | 4 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated |
|---|---|---|---|---|---|---|---|
| Head | Human | 390 | MR, RTDOSE, RTPLAN, RTSTRUCT, CT, REG | Various, Non-Cancer | Limited, Complete | 2022/07/22 |
Summary
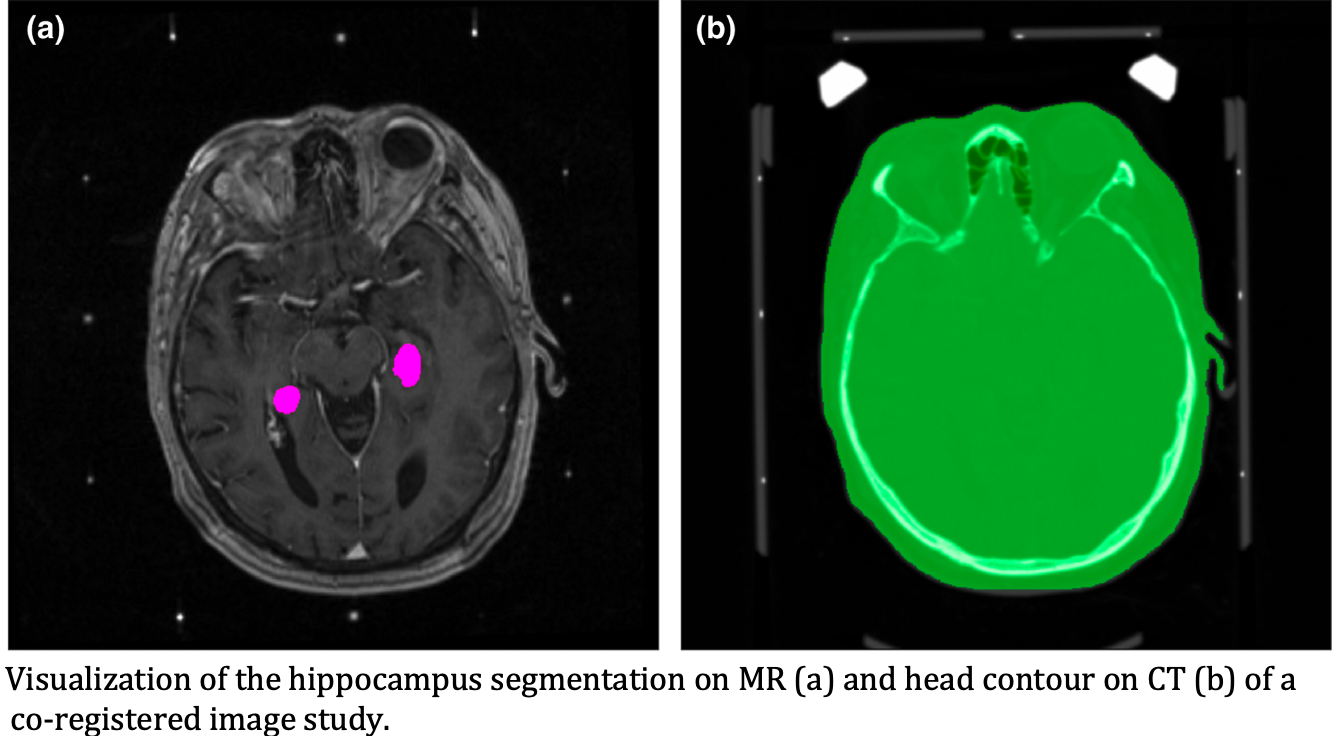
This collection is comprised of 390 patients who presented with vestibular schwannoma (VS, n=73), trigeminal neuralgia (TGN, n=119) or metastatic disease (M, n=198, 4-26 metastases) and were subsequently treated with Gamma Knife (Elekta AB, Stockholm, Sweden) stereotactic radiosurgery. For each patient, the treatment indication is designated with a suffix on their patient ID (VS, TGN or M). All patients are provided with at least one high-resolution (1 mm slice thickness) T1 FLASH trans-axial MR imaging study and their corresponding high-resolution axial planning CT. When available, treatment planning data (struct, dose, plan), alternative MR sequences (FLAIR, T2 CISS, etc.) and follow-up MR imaging studies were collected. Each MR image used during treatment planning was registered to the CT frame of reference and is provided with the DICOM registration file and the aligned secondary image. Additionally, for each patient in the cohort, hippocampal contours generated by multiple institutional observers are provided in a separate structure set. In total, this dataset contains: CT (n=390), MR (n=3868), REG (n=872), DOSE (n=928), PLAN (n=928), planning STRUCT (n=931), and hippocampal research STRUCT (n=390). Gamma Knife planning studies are designated by “Gamma Knife Planning Data” in the study description DICOM header tag. For each patient, MIM (MIM Software, Beachwood, OH) was used to generate a rigid registration between CT and each MR sequence and the registration accuracy was validated using the stereotactic frame fiducial markers. From each registration, a DICOM RTREG file and aligned secondary are provided with each aligned secondaries series indicated by “[original series description] Co-registered to CT” in the series description DICOM header tag. During export from GammaPlan (Elekta AB, Stockholm, Sweden), the treatment planning files are provided in duplicate for each imaging modality frame of reference (CT and each MR sequence). Treatment planning structure names were renamed to remove misspellings, synonyms, and identifiable information. Structures with the format Plan1[tgt#]#Gy denote each target volume (a, b, ...) and the dose to the specified target in Gray. For patients with multiple lesions, ROI structure names are given as the brain region followed by the relative anatomical directions (e.g. Lobe_Frontal_R, Lobe_Parietal_L). Any empty contours were preserved but are designated with '(Empty)' in the name. When available, patient follow-up images were collected between the provided treatment date and either the next Gamma Knife treatment or up-to approximately two years following treatment. In total, 197 patients are provided with follow-up imaging studies, with a median of 2 (range 1-13) follow-up studies provided per patient. Each follow-up imaging study is designated as “Follow-up Image Set #” in the Study Description. Each follow-up study exists in a unique frame of reference and was not co-registered to the original treatment planning CT volume. Three independent observers generated hippocampal contours from the CT aligned-secondary of the T1-weighted MR image, with the resultant contours saved to the CT frame of reference. In total, 744 unique left, right contour pairs were generated (observer 1, n=390; observer 2, n=247; observer 3, n=107). In addition to hippocampal contours, the region grow tool was used to generate a head contour (ROI name ‘head’) to mask out the stereotactic frame and remove most of the reconstruction artifacts on the inferior extent of the image volume. All MR images designated as ‘Co-registered to CT’ in the Series Description were interpolated to the CT frame of reference, potentially altering the field of view or voxel resolution. For all co-registered MR images, the original primary images and registration files are provided for use as needed.Planning Data
Follow-up Data
Hippocampal Contours
Additional Information
Data Access
Some data in this collection contains images that could potentially be used to reconstruct a human face. To safeguard the privacy of participants, users must sign and submit a TCIA Restricted License Agreement to help@cancerimagingarchive.net before accessing the data.
Version 1: Updated 2022/07/22
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images and Radiation Therapy Structures/Doses/Plans | MR, RTDOSE, RTPLAN, RTSTRUCT, CT, REG | DICOM | Download requires NBIA Data Retriever |
390 | 1,509 | 8,307 | 565,921 | TCIA Restricted | View |
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Porter, E., Fuentes, P., Sala, I., Siddiqui, Z., Levitin, R., Myziuk, N., Squires, B., Gonzalez, T., Chen, P., Guerrero, T., & Grills, I. (2022). Gamma Knife MR/CT/RTSTRUCT Sets With Hippocampal Contours (GammaKnife-Hippocampal) (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/Q967-X166 |
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Porter, E., Fuentes, P., Siddiqui, Z., Thompson, A., Levitin, R., Solis, D., Myziuk, N., & Guerrero, T. (2020). Hippocampus segmentation on noncontrast CT using deep learning. Medical Physics, 47(7), 2950–2961. https://doi.org/10.1002/mp.14098 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
