HistologyHSI-GB | Hyperspectral Histological Images for Diagnosis of Human Glioblastoma
DOI: 10.7937/z1k6-vd17 | Data Citation Required | 2.1k Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Brain | Human | 13 | Histopathology, Classification | Glioblastoma | Software/Source Code | Public, Complete | 2024/05/24 |
Summary
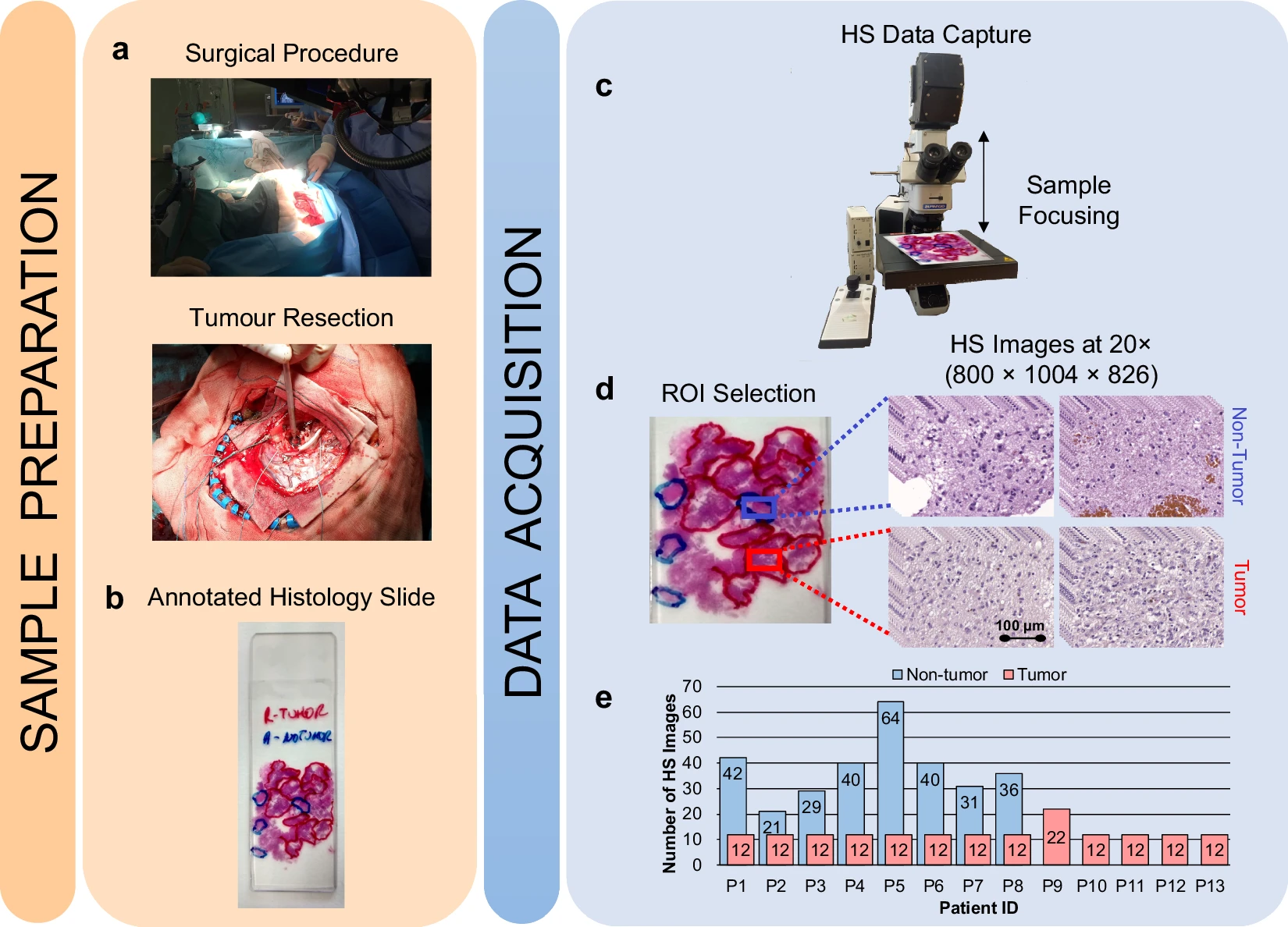
Hyperspectral imaging technology combines the main features of two existing technologies: conventional imaging and spectroscopy. Thus, hyperspectral cameras make it possible to analyze, at the same time and in a non-contact way, the morphological features and chemical composition of the objects captured. The information provided by hyperspectral imaging can be used to detect patterns, cells, or biomarkers to identify diseases. There are different alternatives for processing them and there is a lack of publicly available datasets of medical hyperspectral images. To the best of our knowledge, this is the first open access dataset containing histological hyperspectral images of glioblastoma brain tumors, which can be set as a benchmark for researchers to compare their approaches. This dataset includes 13 subjects. Each subject has a single histological slide with multiple hyperspectral images captured from each slide where deemed relevant by the pathologists (this number varies for each slide). The database is composed of 469 annotated hyperspectral images from 13 histological slides (482 total images), having a spatial dimension of 800 × 1004 pixels and a spectral dimension of 826 spectral channels. The format of the hyperspectral images is ENVI, the standard format for the storage of hyperspectral images. The ENVI format consists of a flat-binary raster file which may or may not have a file extension, accompanied by an ASCII header file (denoted as *.hdr). The data are stored in band-interleaved-by-line format. In addition, dark and white references were captured to perform a calibration of the raw image, which is a standard procedure in hyperspectral image processing. The slides were stained with hematoxylin and eosin and captured using a custom hyperspectral microscopic system at 20× magnification. The ground-truth annotation for this dataset is the diagnosis of the slides (tumor _T_ or not tumor _NT_ ) performed by skilled histopathologists after the visual examination of the stained slides, according to the World Health Organization classification of tumors of the nervous system. As far as we are concerned, there are no commercial hyperspectral whole slide scanners. Also, the availability of hyperspectral microscopes is still limited in the market. The microscope is an Olympus BX-53 (Olympus, Tokyo, Japan). The hyperspectral camera is a Hyperspec® VNIR A-Series from HeadWall Photonics (Fitchburg, MA, USA), which is based on an imaging spectrometer coupled to a charge-coupled device sensor, the Adimec-1000m (Adimec, Eindhoven, Netherlands). This hyperspectral system works in the visual and near-infrared spectral range from 400 to 1000 nm with a spectral resolution of 2.8 nm, sampling 826 spectral channels, and 1004 spatial pixels. The push-broom camera performs a spatial scanning to acquire a hyperspectral cube with a mechanical stage (SCAN, Märzhäuser, Wetzlar, Germany) attached to the microscope, which provides an accurate movement of the slides. The objective lenses are from the LMPLFLN family (Olympus, Tokyo, Japan), optimized for infrared observations. More information about the dataset can be found in this manuscript.
Data Access
Version 1: Updated 2024/05/24
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Tissue Slide Images | Histopathology, Classification | PNG and ENVI | Download requires IBM-Aspera-Connect plugin |
13 | 482 | CC BY 4.0 | — |
Additional Resources for this Dataset
The following external resources have been made available by the data submitters. These are not hosted or supported by TCIA, but may be useful to researchers utilizing this collection.
- A tutorial on how to read and display the hyperspectral data using Python or MATLAB is available at https://github.com/HIRIS-Lab/HistologyHSI-GB.
- The authors also recommend GLIMPS (https://www.rese-apps.com/software/download/form.html) for the visualization of ENVI images.
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Ortega, S., Fabelo, H., Quintana-Quintana, L., Leon, R., Plaza, M.d.l.L., Camacho, R., & Callico, G. M. (2024). Hyperspectral Histological Images for Diagnosis of Human Glioblastoma (HistologyHSI-GB) (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/Z1K6-VD17
|
Acknowledgements
- This work was supported by the Spanish Government and European Union as part of the TALENT-HExPERIA (HypErsPEctRal Imaging for Artificial intelligence applications) project (PID2020-116417RB-C42). Moreover, this work was completed while Laura Quintana and Raquel Leon were beneficiary of the pre-doctoral grant given by the “Agencia Canaria de Investigacion, Innovacion y Sociedad de la Información (ACIISI)” of the “Consejería de Economía, Conocimiento y Empleo”, which is part-financed by the European Social Fund (FSE) (POC 2014-2020, Eje 3 Tema Prioritario 74 (85%)) and, Himar Fabelo was beneficiary of the FJC2020-043474-I funded by MCIN/AEI/10.13039/501100011033 and by the European Union “NextGenerationEU/PRTR”.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Ortega, S., Quintana-Quintana, L., Leon, R., Fabelo, H., Plaza, M. de la L., Camacho, R., & Callico, G. M. (2024). Histological Hyperspectral Glioblastoma Dataset (HistologyHSI-GB). In Scientific Data (Vol. 11, Issue 1). Springer Science and Business Media LLC. https://doi.org/10.1038/s41597-024-03510-x |
Publication Citation |
|
|
Ortega, S., Fabelo, H., Halicek, M., Camacho, R., Plaza, M. de la L., Callicó, G. M., & Fei, B. (2020). Hyperspectral Superpixel-Wise Glioblastoma Tumor Detection in Histological Samples. In Applied Sciences (Vol. 10, Issue 13, p. 4448). MDPI AG. https://doi.org/10.3390/app10134448 |
Publication Citation |
|
|
Ortega, S.; Halicek, M.; Fabelo, H.; Camacho, R.; Plaza, M.d.l.L.; Godtliebsen, F.; M. Callicó, G.; Fei, B. Hyperspectral Imaging for the Detection of Glioblastoma Tumor Cells in H&E Slides Using Convolutional Neural Networks. Sensors 2020, 20, 1911. https://doi.org/10.3390/s20071911 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.