PDMR-Texture-Analysis | Serial Non-contrast Non-gated T2w MRI Datasets of Patient-derived Xenograft Cancer Models for Development of Tissue Characterization Algorithms
DOI: 10.7937/3KQ0-YK19 | Data Citation Required | 932 Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated |
|---|---|---|---|---|---|---|---|
| Arm, Bladder, Buttock, Colon, Liver, Myometrium, Pancreas, Rectum, Shoulder, and Scapula | Mouse | 175 | MR, SR, Protocol, Demographic, Follow-Up, Diagnosis, Other | Ewing sarcoma - Peripheral PNET, Melanoma, Neuroendocrine cancer (NOS), Osteosarcoma, Anal Squamous Cell Carcinoma, Urothelial - bladder cancer (NOS), Pancreatic Adenocarcinoma, Rectal Adenocarcinoma, Colon adenocarcinoma, Lung Squamous Cell Carcinoma | Public, Complete | 2023/06/14 |
Summary
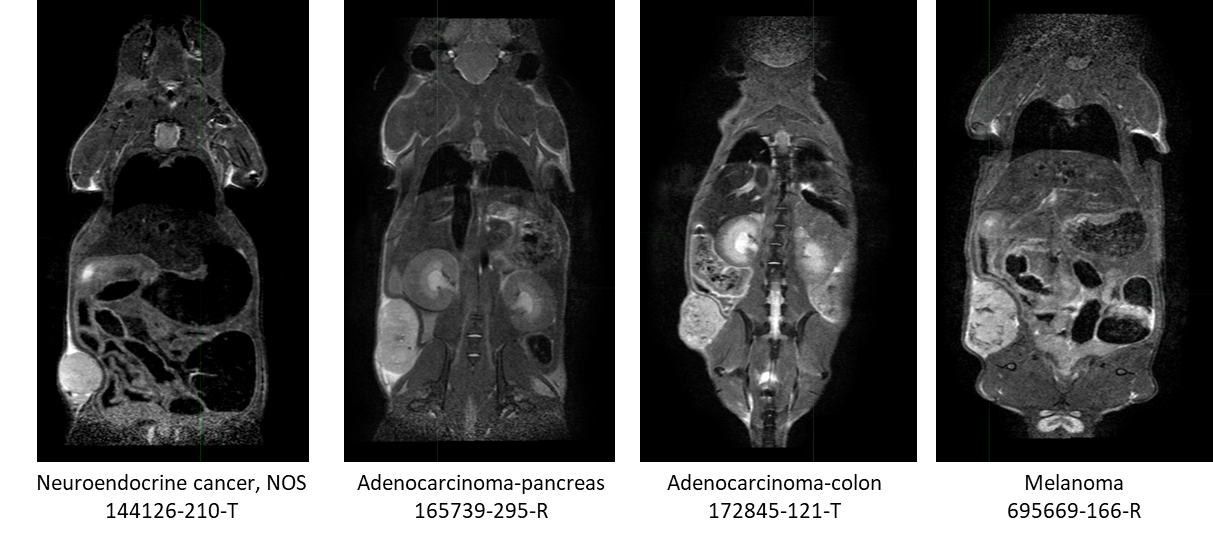
This collection contains serial non-contrast non-gated T2w MRI of 18 patient derived xenograft cancer models. 175 mice were imaged at multiple time points (514 total studies) for researchers to develop algorithms using neural networks, and classification techniques to improve tissue characterization (morphological changes) for the improvement in patient care through advances in precision medicine. Characterization of tissue using non-invasive in vivo imaging techniques is used for detection and measurement of disease burden in oncology. Researchers have developed numerous algorithms, such as neural networks, and classification techniques to improve the characterization (morphological changes) of tissue. Unfortunately, to obtain statistical significance, large datasets are a requirement in this research endeavor due to tumor heterogeneity within the same histologic classification. Pre-clinical patient derived xenograft animal models can be a significant resource by providing collections with a more homogenous tumor genome across the collection with companion genomic and pathologic characterization available (https://pdmr.cancer.gov/), allowing determination of the variability of imaging characteristics. This dataset of a patient derived xenograft model (below table) can be used for training algorithms for evaluating variations in tissue texture with respect to tumor growth and cancer model. Characterization 1 2 3 4 5 6 7 8 Model ID CTEP SDC Description Disease Body Location Biopsy site Implant Date Passage Sex # Mice imaged per biweekly imaging session Neuroendocrine cancer, NOS Endocrine and Neuroendocrine * Liver 2/14/2020 4 M 8 8 5 5 5 5 Urothelial/bladder cancer, NOS Genitourinary Bladder 2/3/2020 4 M 17 16 13 10 11 4 4 Adenocarcinoma-pancreas Digestive/Gastrointestinal Pancreas 5/4/2018 2 M 10 10 1 Adenocarcinoma-colon Digestive/Gastrointestinal * Liver 10/16/2020 4 F 20 20 Adenocarcinoma-colon Digestive/Gastrointestinal * Liver 8/24/2018 3 F 15 13 8 3 Ewing sarcoma/Peripheral PNET Musculoskeletal * Pelvis 3/18/2021 6 M 10 8 1 1 1 Adenocarcinoma-pancreas Digestive/Gastrointestinal Pancreas 12/15/2017 N/A M 5 4 2 1 Adenocarcinoma-pancreas Digestive/Gastrointestinal * Tumor in colonic fat 9/30/2021 4 F 10 10 10 8 5 1 1 Adenocarcinoma-pancreas Digestive/Gastrointestinal * Myometrium 3/27/2018 N/A F 7 7 4 Adenocarcinoma-colon Digestive/Gastrointestinal * Shoulder 8/27/2019 2 F 9 1 Melanoma Skin Arm 4/16/2021 3 M 7 8 8 6 4 4 2 2 Osteosarcoma Musculoskeletal Scapula 3/5/2021 6 F 7 4 Squamous cell lung carcinoma Respiratory/Thoracic * Liver 3/26/2021 4 F 7 8 5 3 1 Adenocarcinoma-rectum Digestive/Gastrointestinal Rectum 2/19/2020 5 F 5 5 5 5 3 4 Adenocarcinoma-pancreas Digestive/Gastrointestinal Pancreas 10/23/2019 2 F 12 12 11 7 Squamous cell carcinoma-anus Digestive/Gastrointestinal Buttock 2/25/2022 5 F 6 6 6 1 1 Adenocarcinoma-colon Digestive/Gastrointestinal * Liver 10/25/2018 3 M 9 2 Urothelial/bladder cancer, NOS Genitourinary Bladder 5/20/2020 4 F 9 6 4 1 1 Note: Biopsy sites labeled with an (*) were obtained from a metastatic site. All other biopsy sites were at the primary tumor site. In this study we performed non-contrast non-gated T2w MRI (SOP50101_MRI), initiated 2 weeks post implantation, and continued biweekly imaging sessions until their tumors reached a size requiring humane termination (ACUC guidance > 2 cm in any linear dimension by caliper or MRI measurement) or their clinical status required euthanasia. Fragments (2x2x2 mm3) from the NCI/DCTD PDMR repository were implanted into 5-10 donor mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG)). When tumors reached enrollment criteria (100 – 300 mm3), tumors were excised, cut into 2x2x2 mm3 fragments and implanted with Matrigel (per PDMR SOP50101_Tumor Implantation) into NSG study mice. The multi-mouse non-gated DICOM dataset was split according to the method published in Tomography and retained their individual mouse DICOM header information. Structured Reports (SR) were added to the dataset to include fragment implant date, CTEP description, mouse strain (NSG) and model. The genomic and pathologic characteristics of these models, which is available from the National Cancer Institute Patient-Derived Models Repository (https://pdmr.cancer.gov/), can be used in conjunction with this publicly available dataset to guide the development of algorithms for enhanced characterization of tissue for precision medicine.PDX Model Characterizations and Biweekly imaging sessions
Data Access
Version 1: Updated 2023/06/14
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images (Full collection — Consult the table in the Summary for Model ID subsets) | MR, SR | DICOM | Download requires NBIA Data Retriever |
175 | 689 | 1,203 | 19,343 | CC BY 4.0 | View |
| Standard Operating Procedure 50101: MRI T2 Weighted Non-Contrast Protocol Single Mouse Pulmonary Gated and Multi-Mouse Non-Gated | Protocol | CC BY 4.0 | — | ||||||
| Standard Operating Procedure 50101: Tumor_Implantation_PDX | Protocol | CC BY 4.0 | — | ||||||
| PDX Model Characterizations | Demographic, Follow-Up, Diagnosis, Other | XLSX | CC BY 4.0 | — | |||||
| Data related to specific models in the collection at NCI Patient-Derived Models Repository PDMR | Diagnosis | DOCX | CC BY 4.0 | — |
Additional Resources for this Dataset
The National Cancer Institute (NCI) has developed a national repository of Patient-Derived Models (PDMs) comprised of patient-derived xenografts (PDXs), in vitro patient-derived tumor cell cultures (PDCs) and cancer associated fibroblasts (CAFs) as well as patient-derived organoids (PDOrg). These models serve as a resource for public-private partnerships and for academic drug discovery efforts. These PDMs are clinically-annotated with molecular information and made available in the Patient-Derived Model Repository.
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Kalen, J. D., Ileva, L. V., Riffle, L. A., Keita, S., Tatum, J. L., Jacobs, P. M., Sanders, C., James, A., Difilippantonio, S., Thang, L., Hollingshead, M. G., Evrard, Y., Clunie, D. A., Miao, T., Wagner, U., Freymann, J., Kirby, J., & Doroshow, J. H. (2023). Serial Non-contrast Non-gated T2w MRI Datasets of Patient Derived Xenograft Cancer Models for Development of Tissue Characterization Algorithms (PDMR-Texture Analysis) (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/3KQ0-YK19 |
Detailed Description
In addition to images, this collection includes Raw Data Storage SOP Class instances with MR Modality, generated by a Philips MR scanner; this data is not useful to anyone without the proprietary software to interpret it.
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
- Frederick National Laboratory for Cancer Research – Special Thanks to Joseph D. Kalen, PhD, Lilia V. Ileva, MS, Lisa A Riffle, Nimit L Patel, MS, Keita Saito, PhD, Yvonne Evrard, PhD, Justin Smith, Simone Difilippantonio, PhD, Chelsea Sanders, Lai Thang, Ulrike Wagner, Yanling Liu, PhD, John B. Freymann, Justin Kirby and Brenda Fevrier-Sullivan
- Division of Cancer Therapeutics and Diagnosis/National Cancer Institute - James L. Tatum, MD, Paula M Jacobs, PhD, Melinda G. Hollingshead, DVM, and James H. Doroshow, MD
- PixelMed Publishing – Special Thanks to David A. Clunie, MD
- University of Arkansas for Medical Sciences – Special Thanks to Kirk E. Smith
- This project has been funded in whole or in part with Federal funds from the National Cancer Institution, National Institutes of Health, under Contract Number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
