Prostate-Anatomical-Edge-Cases | Stress-Testing Pelvic Autosegmentation Algorithms Using Anatomical Edge Cases
DOI: 10.7937/QSTF-ST65 | Data Citation Required | 632 Views | 1 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated |
|---|---|---|---|---|---|---|---|
| Prostate | Human | 131 | CT, RTSTRUCT | Prostate Cancer | Public, Complete | 2023/05/18 |
Summary
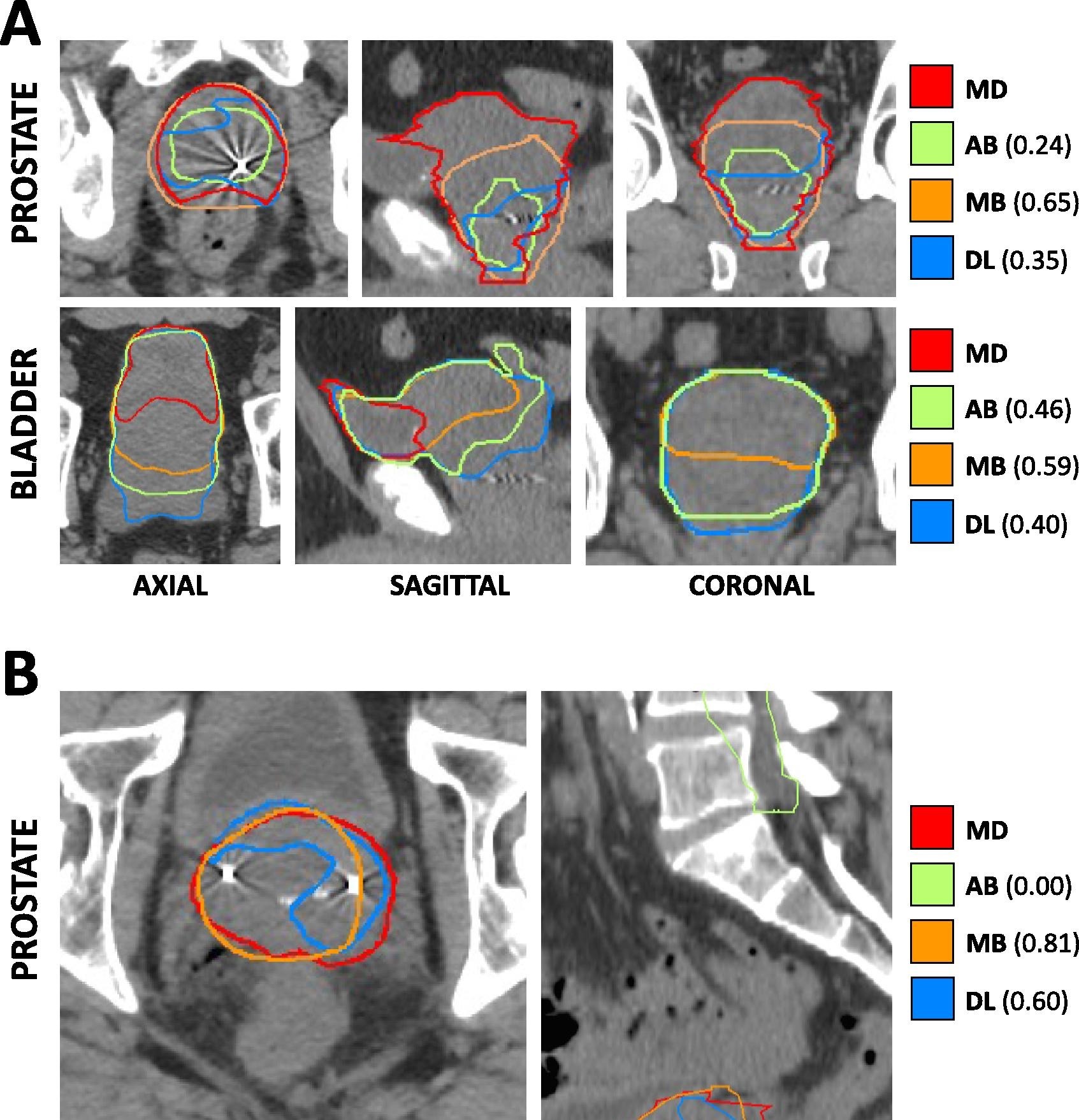
In this single institution retrospective study, we reviewed 950 consecutive patients with prostate adenocarcinoma receiving definitive radiotherapy between 2011 and 2019, and identified among them 112 patients with anatomic variations (edge cases) seen on simulation CT and/or MRI imaging. These variations included hip arthroplasty, prostate median lobe hypertrophy, so-called “droopy” seminal vesicles, presence of a urinary catheter, and others. A separate cohort of 19 “normal” cases were randomly selected for inclusion. Prostate, rectum, bladder, and bilateral femoral heads were manually segmented on all CT simulation images (where present) and were ultimately used clinically for radiation treatment planning. We leveraged this imaging dataset to assess the comparative performance of deep learning, atlas-based, and model-based autosegmentation methods across both normal and edge case cohorts: https://doi.org/10.1016/j.phro.2023.100413. In this paper and in the figure on the right, we show the Cross-sectional CT-based anatomy and autosegmentation performance for representative edge cases. A) Hypertrophic prostate edge case. Each panel depicts a focused excerpt from a single CT scan, centered about two different structures (prostate, bladder) in three different planes (axial, sagittal, coronal). Clinician-delineated “ground truth” contours (MD) for each structure are shown in red, while atlas-based (AB), model-based (MB), and deep-learning based (DL) autosegmented contours are depicted in green, orange, and blue, respectively. Numerical values represent DSC for the corresponding autosegmented volumes compared to MD volumes. B) So-called “droopy” seminal vesicles edge case. Each panel depicts a focused excerpt from a single CT scan, centered about the prostate in two different planes (axial, sagittal). All colors and labeling are as in Panel A).
Data Access
Version 1: Updated 2023/05/18
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images and Radiation Therapy Structures | CT, RTSTRUCT | DICOM | Download requires NBIA Data Retriever |
131 | 131 | 262 | 23,490 | CC BY 4.0 | View |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Thompson, R. F., Kanwar, A., Merz, B., Cohen, E., Fisher, H., Rana, S., Claunch, C., & Hung, A. (2023). Stress-Testing Pelvic Autosegmentation Algorithms Using Anatomical Edge Cases (Prostate Anatomical Edge Cases) (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/QSTF-ST65 |
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Kanwar, A., Merz, B., Claunch, C., Rana, S., Hung, A., & Thompson, R. F. (2023). Stress-testing pelvic autosegmentation algorithms using anatomical edge cases. In Physics and Imaging in Radiation Oncology (Vol. 25, p. 100413). Elsevier BV. https://doi.org/10.1016/j.phro.2023.100413 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
