Radiomics-Tumor-Phenotypes | Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach
DOI: 10.7937/K9/TCIA.2014..UA0JGPDG | Data Citation Required | 424 Views | 4 Citations | Analysis Result
| Location | Subjects | Size | Updated | |||
|---|---|---|---|---|---|---|
| Lung Cancer, Head and Neck Cancer | Lung, Head-Neck | 1,019 | Genomics | 2020/03/23 |
Summary
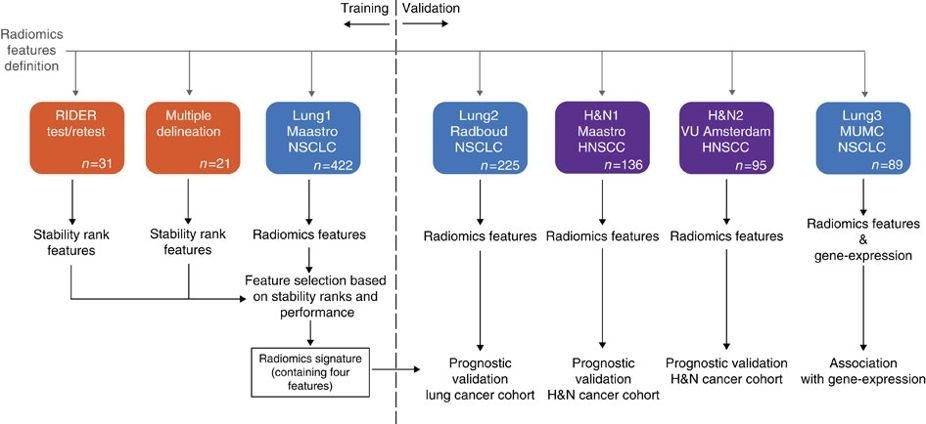
This data applies a radiomic approach to computed tomography data of 1,019 patients with lung or head-and-neck cancer which are described in Nature Communications (http://doi.org/10.1038/ncomms5006). The various arms of the study are represented in TCIA as distinct Collections including NSCLC-Radiomics (Lung1), NSCLC-Radiomics-Genomics (Lung3), Head-Neck-Radiomics-HN1 (H&N1), NSCLC-Radiomics-Interobserver1 (Multiple delineation), and RIDER-LungCT-Seg (RIDER test/retest). Radiomics refers to the comprehensive quantification of tumour phenotypes by applying a large number of quantitative image features. In present analysis 440 features quantifying tumour image intensity, shape and texture, were extracted. We found that a large number of radiomic features have prognostic power in independent data sets, many of which were not identified as significant before. Radiogenomics analysis revealed that a prognostic radiomic signature, capturing intra-tumour heterogeneity, was associated with underlying gene-expression patterns. These data suggest that radiomics identifies a general prognostic phenotype existing in both lung and head-and-neck cancer. This may have a clinical impact as imaging is routinely used in clinical practice, providing an unprecedented opportunity to improve decision-support in cancer treatment at low cost.
Data Access
Version 2: Updated 2020/03/23
Added links to the recently published TCIA collections which reflect the additional arms of the study described in Nature Communications (http://doi.org/10.1038/ncomms5006).
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Gross Tumor Volume Segmentations from RIDER-LungCT-Seg | RTSTRUCT, SEG | DICOM | Download requires NBIA Data Retriever |
31 | 43 | 118 | 118 | CC BY 3.0 | View |
Collections Used In This Analysis Result
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Corresponding Original Images from Head-Neck-Radiomics-HN1 - H&N1 | CT, SEG, RTSTRUCT, PT | DICOM | Requires NBIA Data Retriever |
137 | 137 | 486 | 28,918 | TCIA No Commercial Limited | View |
| Corresponding Original Images from NSCLC-Radiomics, NSCLC-Radiomics-Genomics, NSCLC-Radiomics-Interobserver1 | CT, SEG, RTSTRUCT | DICOM | Requires NBIA Data Retriever |
533 | 533 | 1,418 | 69,441 | CC BY-NC 3.0 | View |
| Corresponding Original Images from RIDER-LungCT-Seg - RIDER test/retest | RTSTRUCT, SEG | DICOM | Requires NBIA Data Retriever |
31 | 43 | 118 | 118 | CC BY 3.0 | View |
Additional Resources For This Dataset
The following external resources have been made available by the data submitters. These are not hosted or supported by TCIA, but may be useful to researchers utilizing this collection.
- Genomics data in Gene Expression Omnibus for NSCLC-Radiomics-Genomics (Lung3) Gene Expression Data
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Aerts, H., Velazquez, E. R., Leijenaar, R. T. H., Parmar, C., Grossmann, P., Carvalho, S., Bussink, J., Monshouwer, R., Haibe-Kains, B., Rietveld, D., Hoebers, F., Rietbergen, M. M., Leemans, C. R., Dekker, A., Quackenbush, J., Gillies, R. J., & Lambin, P. (2014). Data from: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach (Radiomics-Tumor-Phenotypes). [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2014..UA0JGPDG |
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Aerts, H. J. W. L., Velazquez, E. R., Leijenaar, R. T. H., Parmar, C., Grossmann, P., Carvalho, S., Bussink, J., Monshouwer, R., Haibe-Kains, B., Rietveld, D., Hoebers, F., Rietbergen, M. M., Leemans, C. R., Dekker, A., Quackenbush, J., Gillies, R. J., & Lambin, P. (2014). Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Communications, 5(1). https://doi.org/10.1038/ncomms5006 |
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
