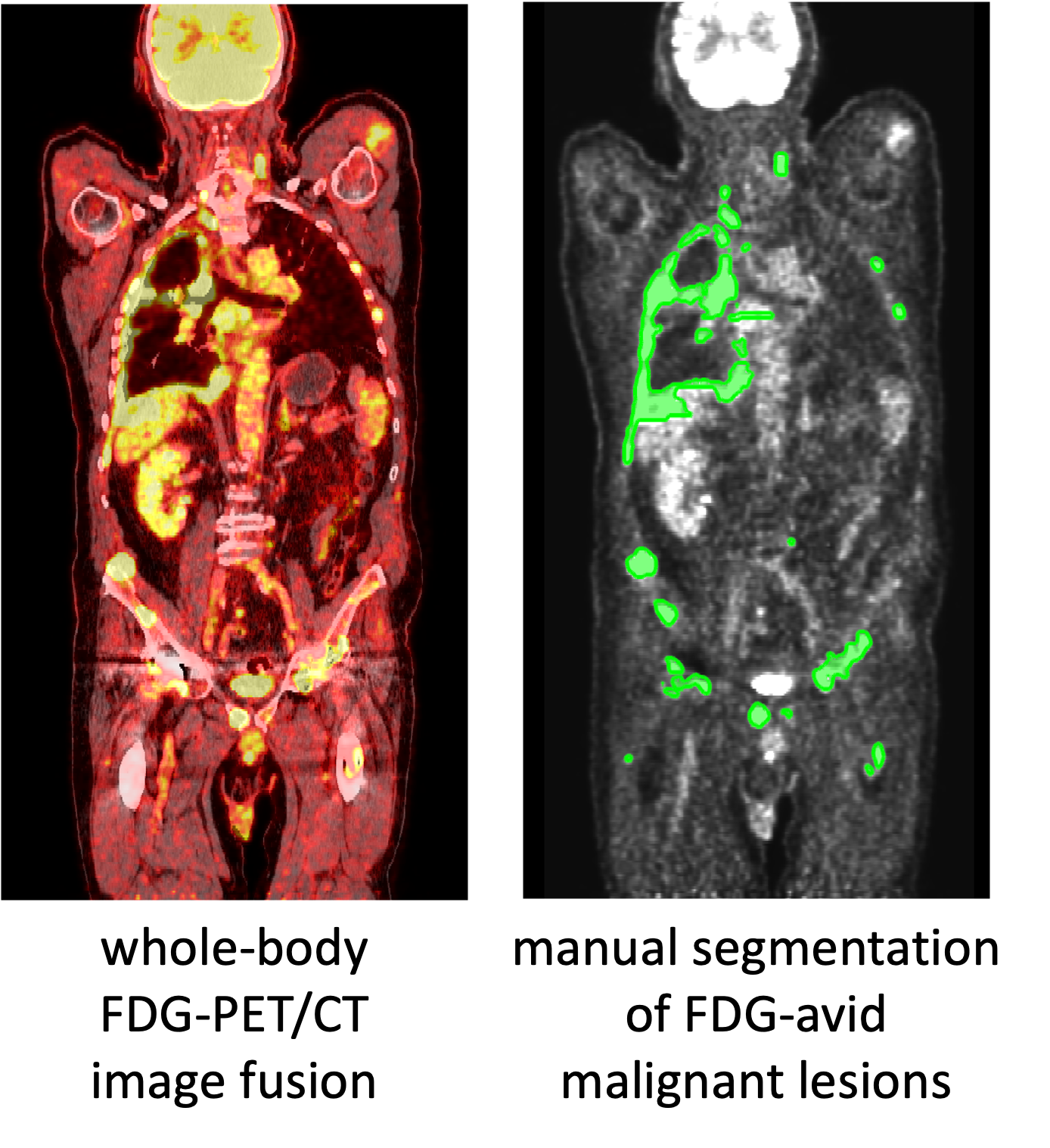
FDG-PET-CT-Lesions | A whole-body FDG-PET/CT dataset with manually annotated tumor lesions
DOI: 10.7937/gkr0-xv29 | Data Citation Required | 10.6k Views | 14 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Lung, Lymph, and Skin | Human | 900 | CT, PT, SEG, Demographic, Diagnosis | Lymphoma, Melanoma, Non-small Cell Lung Cancer | Clinical, Image Analyses, Software/Source Code | Limited, Complete | 2022/06/02 |
Summary
Purpose: To provide an annotated data set of oncologic PET/CT studies for the development and training of machine learning methods and to help address the limited availability of publicly available high-quality training data for PET/CT image analysis projects. This data can also be used for machine learning challenges, which is exemplified in the autoPET MICCAI 2022 competition: https://autopet.grand-challenge.org/. Data: The anonymized publication of data was approved by the local ethics committee and data protection officer. 501 consecutive whole body FDG-PET/CT data sets of patients with malignant lymphoma, melanoma and non small cell lung cancer (NSCLC) as well as 513 data sets without PET-positive malignant lesions (negative controls) examined between 2014 and 2018 at the University Hospital Tübingen were included. All examinations were acquired on a single, state-of-the-art PET/CT scanner (Siemens Biograph mCT). The imaging protocol consists of a diagnostic CT scan (mainly from skull base to mid-thigh level) with intravenous contrast enhancement in most cases, except for patients with contraindications. The following CT parameters were used: reference dose of 200 mAs, tube voltage of 120 kV, iterative reconstruction with a slice thickness of 2 - 3 mm. In addition, a whole-body FDG-PET scan was acquired 60 minutes after I.V. injection of 300-350 MBq 18F-FDG. PET data were reconstructed using an ordered-subset expectation maximization (OSEM) algorithm with 21 subsets and 2 iterations and a gaussian kernel of 2 mm and a matrix size of 400 x 400. All data sets were analyzed in a clinical setting by a radiologist and nuclear medicine physician in consensus identifying primary tumors and metastases in each data set. All FDG-avid lesions identified as malignant based on patient history and prior examinations were manually segmented on PET images in a slice-per-slice manner by a single reader using dedicated software (NORA imaging platform, University of Freiburg, Germany). We provide the anonymized original DICOM files of all studies as well as the DICOM segmentation masks. Primary diagnosis, age and sex are provided as non-imaging information (csv). In addition, we provide links to code for you to make a preprocessed version of the data with resampled and aligned PET, CT, and masks as a NIfTI file and in the hdf5 format ready to use in machine learning projects.
Data Access
Some data in this collection contains images that could potentially be used to reconstruct a human face. To safeguard the privacy of participants, users must sign and submit a TCIA Restricted License Agreement to help@cancerimagingarchive.net before accessing the data.
Version 1: Updated 2022/06/02
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images | CT, PT, SEG | DICOM | Download requires NBIA Data Retriever |
900 | 1,014 | 3,042 | 916,957 | TCIA Restricted | View |
| Clinical data | Demographic, Diagnosis | CSV | CC BY 4.0 | — |
Additional Resources for this Dataset
The following external resources have been made available by the data submitters. These are not hosted or supported by TCIA, but may be useful to researchers utilizing this collection.
- Scripts provided by the submitting group for file conversion, preprocessing alignment and resampling of PET, CT and mask data to NIfTI, MHA, and HDF5 formats: https://github.com/lab-midas/TCIA_processing
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Gatidis S, Kuestner T. (2022) A whole-body FDG-PET/CT dataset with manually annotated tumor lesions (FDG-PET-CT-Lesions) [Dataset]. The Cancer Imaging Archive. DOI: 10.7937/gkr0-xv29 |
Detailed Description
Notes:
Here are conversion scripts for these data https://github.com/lab-midas/TCIA_processing
- Converts DICOM to NIfTI , and also create resampled/resliced CT and an SUV file using tcia_dicom_to_nifti.py (requires install of dicom2nifti and matplotlib)
- It is straight forward to generate HDF5 files from the NIfTI files using tcia_nifti_to_hdf5.py.
- Organizes NIfTI into HDF5 structure; note this output is a single large package.
SEG are most easily reviewed as overlay using MITK viewer or 3D Slicer.
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
- University Hospital Tübingen, Tübingen, Germany - Special thanks to
- Christian La Fougère, MD from the Department of Nuclear Medicine
- Tobias Hepp, MD from the Department of Radiology
- Konstantin Nikolaou, MD from the Department of Radiology
- Christina Pfannenberg, MD from the Department of Radiology
- University Hospital of the LMU (Munich), Germany – Special thanks to
- Clemens Cyran, MD from the Department of Radiology
- Michael Ingrisch from the Department of Radiology
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Gatidis, S., Hepp, T., Früh, M., La Fougère, C., Nikolaou, K., Pfannenberg, C., Schölkopf, B., Küstner, T., Cyran, C., & Rubin, D. (2022). A whole-body FDG-PET/CT Dataset with manually annotated Tumor Lesions. In Scientific Data (Vol. 9, Issue 1). DOI: 10.1038/s41597-022-01718-3 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
