GBM-DSC-MRI-DRO | GBM-DSC-MRI-DRO
DOI: 10.7937/TCIA.2020.RMWVZWIX | Data Citation Required | 345 Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated |
|---|---|---|---|---|---|---|---|
| Brain Phantom | Human | 3 | MR, Other | Phantom | Public, Complete | 2022/07/21 |
Summary
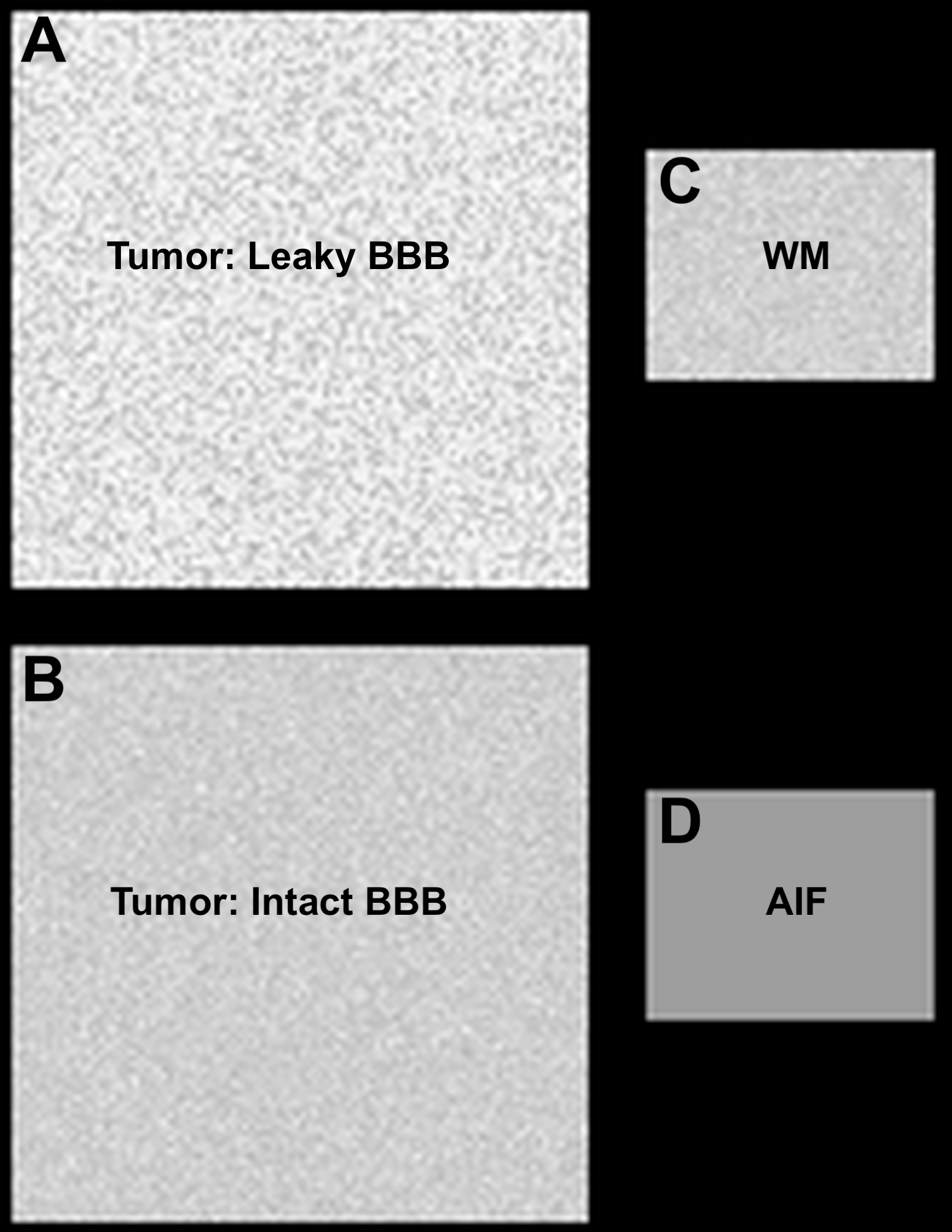
The standardization of dynamic susceptibility contrast (DSC)-magnetic resonance imaging (MRI) has been confounded by a lack of consensus on DSC-MRI methodology for preventing potential relative cerebral blood volume (CBV) inaccuracies, including the choice of acquisition protocols and postprocessing algorithms. Therefore, a digital reference object (DRO) was developed using physiological and kinetic parameters derived from a patient database, unique voxel-wise 3-dimensional tissue structures, and a validated MRI signal computational approach. The primary, intended use of the DRO is to validate image acquisition and analysis methods for accurately measuring relative cerebral blood volume in glioblastomas [1,2]. The DRO datasets have also been used as part of the QIN-challenge titled “DSC-DRO Challenge” to evaluate multisite rCBV consistency [3], and for systematic assessment of multi-echo DSC-MRI [4]. References: To achieve DSC-MRI signals representative of the temporal characteristics, magnitude, and distribution of contrast agent-induced T1 and T2 * changes observed across multiple glioblastomas, the DRO’s input parameters were trained using DSC-MRI data from 23 glioblastomas (40,000 voxels). The DRO’s ability to produce reliable signals across combinations of pulse sequence parameters and contrast agent dosing schemes unlike those in the training data set was validated by comparison with in vivo dual-echo DSC-MRI data acquired in a separate cohort of patients with glioblastomas. To achieve an excellent agreement between the DRO and in vivo data the training and validation process required a DRO consisting of 10 000 unique voxels. Users can use the DRO to investigate the influence of DSC-MRI acquisition and post-processing methods on CBV accuracy and as a benchmark for perfusion analysis algorithms. The DRO data is separated in to two collections corresponding to two magnetic field strengths (3T and 1.5T). Each collection contains 15 folders corresponding to three TRs (1s, 1.5s, 2s) and five contrast agent dosing schemes (pre none bolus full, pre quarter bolus three-quarter, pre half bolus half, pre quarter bolus full, and pre full bolus full). Each of the 15 folders contains 12 sub-folders representing a combination of three flip angles (30o, 60o, and 90o) and four echo times (20ms, 30ms, 40ms, and 50ms). Within each of the 12 sub-folders a single slice DSC-MRI signal time series of 3 minutes sampled at intervals of the corresponding TR values are given. The slice contains four ROIs described in figure 1. In addition to the DRO data masks for each of the four ROIs are provided. Figure 1 (top right of page): (A) Represents the tumor region containing 10000 voxels. (B) The corresponding tumor region with no CA leakage contamination (to be used for expected calculations). (C) Representative normal appearing white matter (WM) containing 2000 voxels. (D) Region representing the arterial input function (AIF). For a given parameter combination (TR=1.5s, TE=30ms, Flip angle= 60o, CA dosing = pre none bolus full, and B0= 3T), Figure 2 demonstrate example signal time course for all four ROI. Figure 2 (below): Example signal time course for a voxel within each of the four ROIs.DRO development
Application
Guide
Data Access
Version 1: Updated 2022/07/21
Added new subjects.
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images | MR | DICOM | Download requires NBIA Data Retriever |
3 | 31 | 364 | 47,164 | CC BY 4.0 | View |
| Data description | Other | CC BY 4.0 | — |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Semmineh, N. B., Stokes, A. M., Bell, L. C., Boxerman, J. L., & Quarles, C. C. (2020). GBM-DSC-MRI-DRO [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.2020.RMWVZWIX |
Detailed Description
The field of view contains four regions of interest (ROI) per image. The settings for
- Static field strength,
- TR,
- TE,
- Flip angle,
- and contrast dose
are included in the series description of each synthetic timecourse, as in Table 1 of the publication .
Note, do not sort by filename (chart at left), sort by Acquisition Number (chart at right). It should trace like the image on the right. Osirix, IBNeuro, ImageJ do this natively. If you are using Matlab or python please use DICOM header AcquisitionNumber (0020,0012) to reorder the files; using the file names could lead to a wrong signal profile.
Acknowledgements
- This work was performed at the Barrow Neurological Institute, with support from R01 CA158079.
- We thank Dr. Kathleen Schmainda (Medical College of Wisconsin) for access to dual-echo data used for validation.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Semmineh, N. B., Stokes, A. M., Bell, L. C., Boxerman, J. L., & Quarles, C. C. (2017). A Population-Based Digital Reference Object (DRO) for Optimizing Dynamic Susceptibility Contrast (DSC)-MRI Methods for Clinical Trials. In Tomography (Vol. 3, Issue 1, pp. 41–49). MDPI AG. https://doi.org/10.18383/j.tom.2016.00286 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.


