PDMR-833975-119-R | Imaging tissue characterization of a patient derived xenograft model of adenocarcinoma pancreas:
DOI: 10.7937/TCIA.0ECK-C338 | Data Citation Required | 200 Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Abdomen | Mouse | 20 | MR, SR, Protocol | Pancreatic Ductal Adenocarcinoma | Clinical | Public, Complete | 2020/10/05 |
Summary
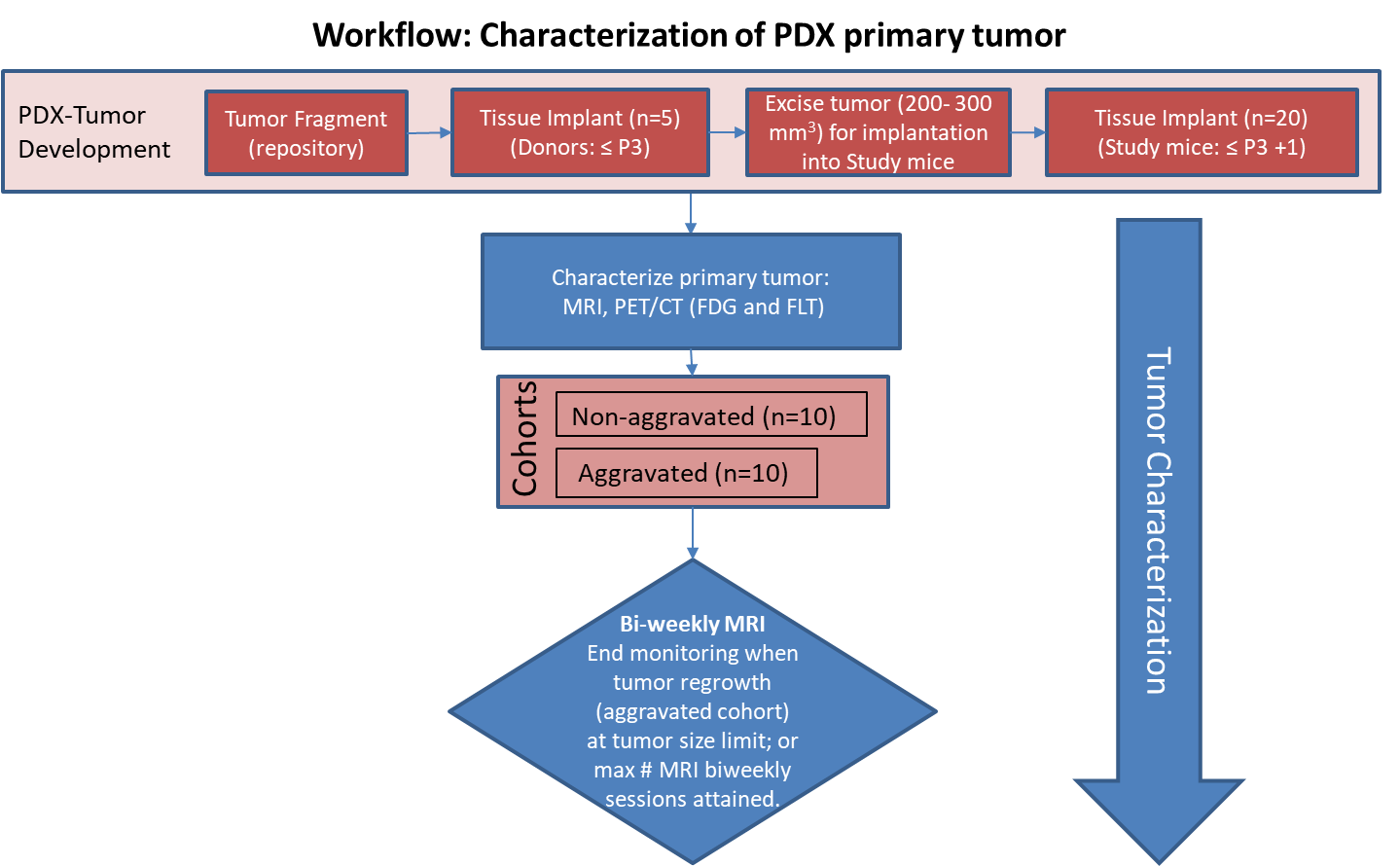
Characterization of tissue using in vivo non-invasive imaging is the foundation of Radiology and is clinically used for detection and measurement of disease burden in oncology. With the migration of imaging to digital media the possibility for advanced mathematically based imaging biomarkers was realized. As part of this endeavor, researchers have developed various algorithms using neural networks, and classification techniques to improve tissue characterization (morphological changes). However, large datasets are a requirement in this research endeavor in part due to the genomic heterogeneity of tumors in the same histologic classification – many tumors from different patients are required to have enough with the same genomic characteristics to adequately evaluate the range of imaging variability for a specific genomic pattern. Pre-clinical animal models of patient derived xenografts may be an important resource by providing collections with a more homogenous tumor genome across the collection with companion extensive tumor genomic characterization available, allowing determination of the variability of imaging characteristics for that pattern in different individuals. This dataset of a patient derived xenograft model adenocarcinoma pancreas PDMR: 833975-119-R can be used for training algorithms for evaluating variations in tissue texture with respect to tumor growth and regrowth after surgical resection. In this study we performed a detailed imaging characterization (workflow below) of this model, details are provided in the attached standard operating procedures. Tumors in half of the mice were resected in the range 200-300 mm3 size; tumors in the other half were allowed to grow until it was necessary to euthanize them because of tumor size. T2w MRI at 56 days post implant demonstrated a heterogenous tumor with an apparent central hemorrhage and the implant appears to have a defined capsule. On day 98 (post-implant) the tumor increased in size (618%) with very similar imaging characteristics as displayed on day 56. In contrast, the regrowth following surgical resection of the primary xenograft (105 days post resection) is homogenous, but also appears to have a well-defined capsule. PET/CT Characterization of the primary tumor: Baseline PET (SOP attached) were performed when tumor reached an approximate 200 mm3. Average SUVbw_max values (n=5) were calculated; [18F]FDG: 1.8 ± 0.2 and [18F]FLT: 2.8 ± 0.7. The imaging characteristics of this model, which is available from the National Cancer Institute Patient-Derived Models Repository (https://pdmr.cancer.gov/), is highly favorable for preclinical research studies when used in conjunction with non-contrast T2 weighted MRI.
Data Access
Version 1: Updated 2020/10/05
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images | MR, SR | DICOM | Download requires NBIA Data Retriever |
20 | 100 | 180 | 2,980 | CC BY 3.0 | View |
| Standard Operating Procedure 50101: MRI T2 Weighted Non-Contrast imaging protocol: Single Mouse Pulmonary Gated and Multi-Mouse Non-Gated | Protocol | CC BY 3.0 | — | ||||||
| Standard Operating Procedure 50102: Positron Emission Tomography PET imaging protocol | Protocol | CC BY 3.0 | — |
Additional Resources for this Dataset
The National Cancer Institute (NCI) has developed a national repository of Patient-Derived Models (PDMs) comprised of patient-derived xenografts (PDXs), in vitro patient-derived tumor cell cultures (PDCs) and cancer associated fibroblasts (CAFs) as well as patient-derived organoids (PDOrg). These models serve as a resource for public-private partnerships and for academic drug discovery efforts. These PDMs are clinically-annotated with molecular information and made available in the Patient-Derived Model Repository. Data related to the specific subjects in this Collection can be found at:
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Tatum, J., Kalen, J.D., Ileva, L.V., Riffle, L.A., Saito, K., Patel, N., Jacobs, P., Sanders, C., James, A., Difilippantonio, S., Thang, L., Hollingshead, M.G., Phillips, J., Evrard, Y., Clunie, D., Liu, Y., Smith, K.E., Wagner, U., Freymann, J.B., Kirby, J., Doroshow, J. (2020). Imaging tissue characterization of a patient derived xenograft model of adenocarcinoma pancreas: (PDMR-833975-119-R) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.0ECK-C338 |
Detailed Description
In addition to images, this collection includes Raw Data Storage SOP Class instances with MR Modality, generated by a Philips MR scanner; this data is not useful to anyone without the proprietary software to interpret it.
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
Frederick National Laboratory for Cancer Research – Special Thanks to Joseph D. Kalen, PhD, Lilia V. Ileva, MS, Lisa A Riffle, Nimit Patel, Keita Saito, PhD, Yvonne Evrard, PhD, Jessica Phillips, Simone Difilippantonio, PhD, Chelsea Sanders, Amy James, Lia Thang, Ulrike Wagner, Yanling Liu, PhD, John B. Freymann, and Justin Kirby.
Division of Cancer Therapeutics and Diagnosis/National Cancer Institute - James L. Tatum, MD, Paula M Jacobs, PhD, Melinda G. Hollingshead, DVM, and James H. Doroshow, MD
PixelMed Publishing – Special Thanks to David A. Clunie, MD
University of Arkansas for Medical Sciences – Special Thanks to Kirk E. Smith
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract Number HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
