Vestibular-Schwannoma-MC-RC | Segmentation of Vestibular Schwannoma from Magnetic Resonance Imaging: An Annotated Multi-Center Routine Clinical Dataset
DOI: 10.7937/HRZH-2N82 | Data Citation Required | 1k Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated |
|---|---|---|---|---|---|---|---|
| Ear | Human | 124 | MR, Segmentation, Other | Vestibular Schwannoma (non-cancer) | Public, Complete | 2023/08/23 |
Summary
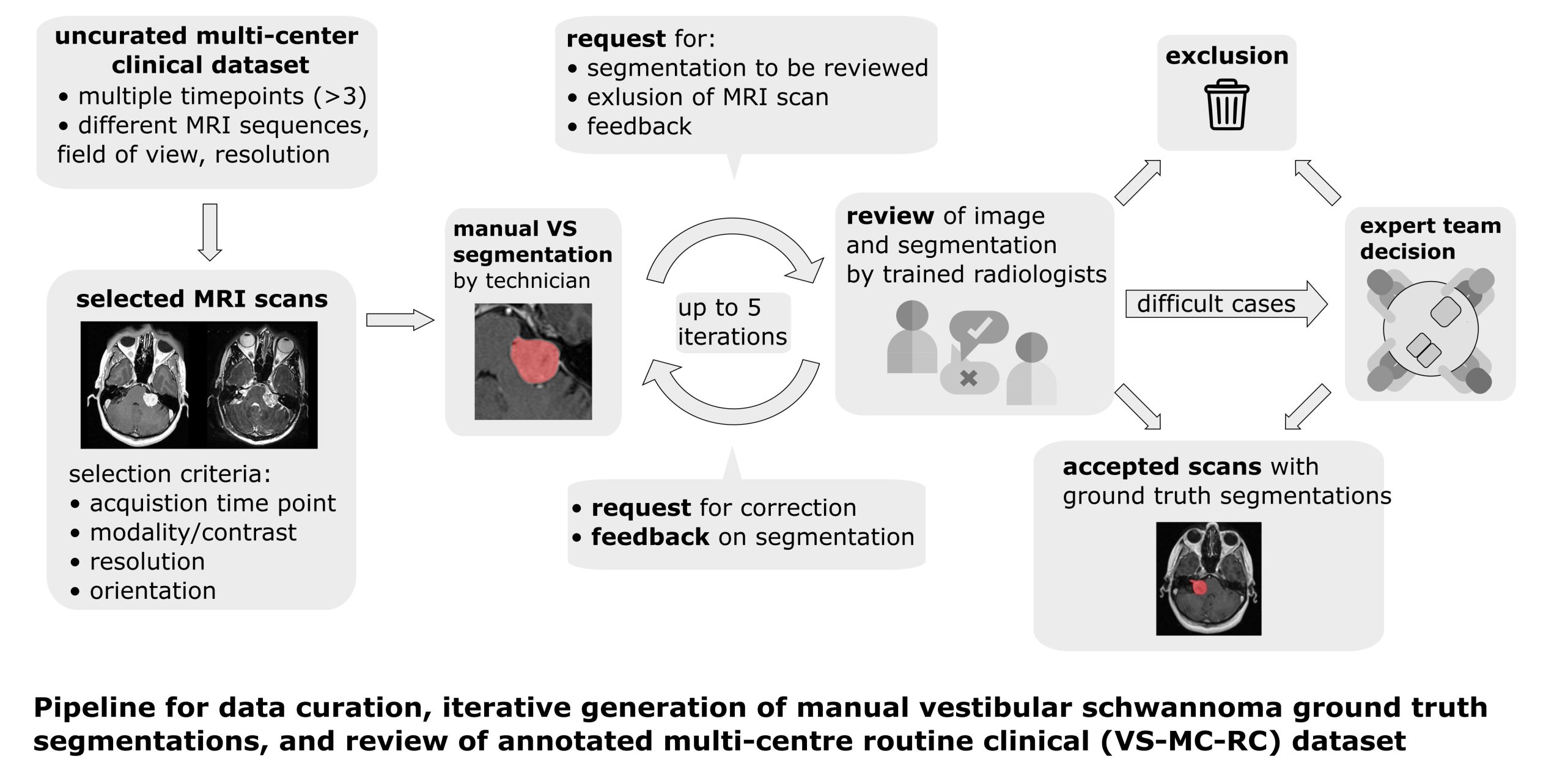
The Vestibular-Schwannoma-MC-RC (VS-MC-RC) dataset contains longitudinal MRI scans of 160 patients with a single sporadic Vestibular Schwannoma (VS) from 10 medical sites in the United Kingdom. For all patients either a T1-weighted (T1w) or T2-weighted (T2w) scan, or both, are available. The dataset provides binary segmentations of the VS for all scan sessions. The dataset comprises up to three time points for each patient resulting in a total of 427 timepoints and 487 3D-images. These timepoints represent the first, central, and last MRI exams that the patients underwent. For 60 timepoints, both T1w and T2w are available, for 64 timepoints only T1w, and for 303 timepoints only T2w. Manual ground truth segmentations were obtained in an iterative process in which segmentations were: 1) produced or amended by a specialized company; and 2) reviewed by one of three trained radiologists; and 3) validated by an expert team. Using this dataset, researchers can develop and validate methods, for example, for automatic surveillance of Vestibular Schwannoma, which work robustly on images acquired at different hospitals. Pre-processing The MRI images are provided in DICOM format. All images were defaced using automatic affine registration or manual landmark registration with BRAINSFit and the template and face mask provided in pydeface. Segmentations are provided in NIfTI format Comparison to VS-SEG dataset Compared to the Vestibular-Schwannoma-SEG (VS-SEG) dataset which was acquired with a standardized protocol for radiation treatment planning on a single scanner at a single site, the VS-MC-RC dataset aims to capture the diversity of MRI scans encountered in a routine clinical setting; it contains a wide range of scan protocols and was acquired on multiple scanners by different manufacturers. In addition, the tumour manifestation in scans of this dataset is more diverse because it includes tumours at early stages and post-operative tumour residuals rather than only tumours specifically selected for radiation therapy. Patient overlap with VS-SEG dataset 58 patients of the VS-MC-RC dataset were previously included in the VS-SEG dataset. These patients have a Patient ID that begins with “VS-SEG- “. Patient IDs of patients that are not part of the VS-SEG dataset begin with “VS-MC-RC-”. Please note that for most patients that are part of both datasets, the timepoints and MRI series included in one dataset are different from those included in the other dataset. However, 8 series are included in both datasets. These series are listed in the CSV file below. Please note that the VS-SEG dataset was not exported with the same software as the VS-MC-RC dataset. Therefore, the metadata of overlapping series might differ. Furthermore, different defacing algorithms were used so that pixel data of overlapping series are not the same in affected areas. Embargoed data This dataset is part of the challenge “Cross-Modality Domain Adaptation for Medical Image Segmentation” (crossMoDA). 126 timepoints that were included in the validation and test set of the challenge are currently under embargo and will be released in August 2024. Until then, 301 timepoints from 124 patients are available for download.
Data Access
Version 1: Updated 2023/08/23
Note: There are 124 patients currently available in TCIA search and download; the remaining 36 patients will be available post challenge in 2024.
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images | MR | DICOM | Download requires NBIA Data Retriever |
124 | 301 | 354 | 22,086 | CC BY 4.0 | View |
| Segmentations | Segmentation | NIFTI and ZIP | CC BY 4.0 | — | |||||
| Mapping of series in Vestibular-Schwannoma-SEG dataset | Other | CSV | CC BY 4.0 | — |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Kujawa, A., Dorent, R., Wijethilake, N., Connor, S., Thomson, S., Ivory, M., Bradford, R., Kitchen, N., Bisdas, S., Ourselin, S., Vercauteren, T., & Shapey, J. (2023). Segmentation of Vestibular Schwannoma from Magnetic Resonance Imaging: An Annotated Multi-Center Routine Clinical Dataset (Vestibular-Schwannoma-MC-RC) (Version 1) [dataset]. The Cancer Imaging Archive. https://doi.org/10.7937/HRZH-2N82 |
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
- This work was supported by Wellcome Trust (203145Z/16/Z, 203148/Z/16/Z, WT106882), EPSRC (NS/A000050/1, NS/A000049/1) and MRC (MC/PC/180520) funding.
- Additional funding was provided by Medtronic.
- Tom Vercauteren is also supported by a Medtronic/Royal Academy of Engineering Research Chair (RCSRF1819/7/34).
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Kujawa, A., Dorent, R., Connor, S., Thomson, S., Ivory, M., Vahedi, A., Guilhem, E., Wijethilake, N., Bradford, R., Kitchen, N., Bisdas, S., Ourselin, S., Vercauteren, T., & Shapey, J. (2024). Deep learning for automatic segmentation of vestibular schwannoma: a retrospective study from multi-center routine MRI. In Frontiers in Computational Neuroscience (Vol. 18). Frontiers Media SA. https://doi.org/10.3389/fncom.2024.1365727 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
