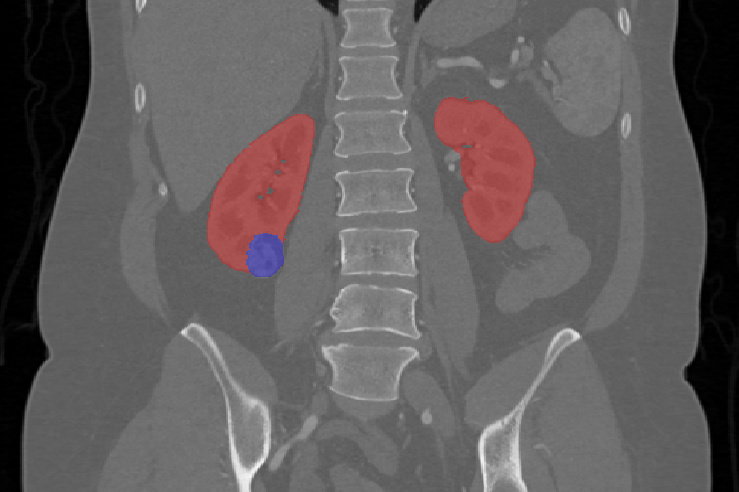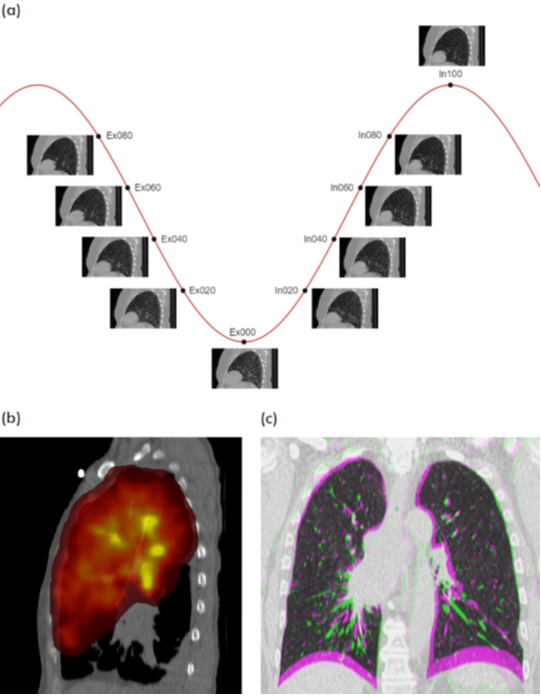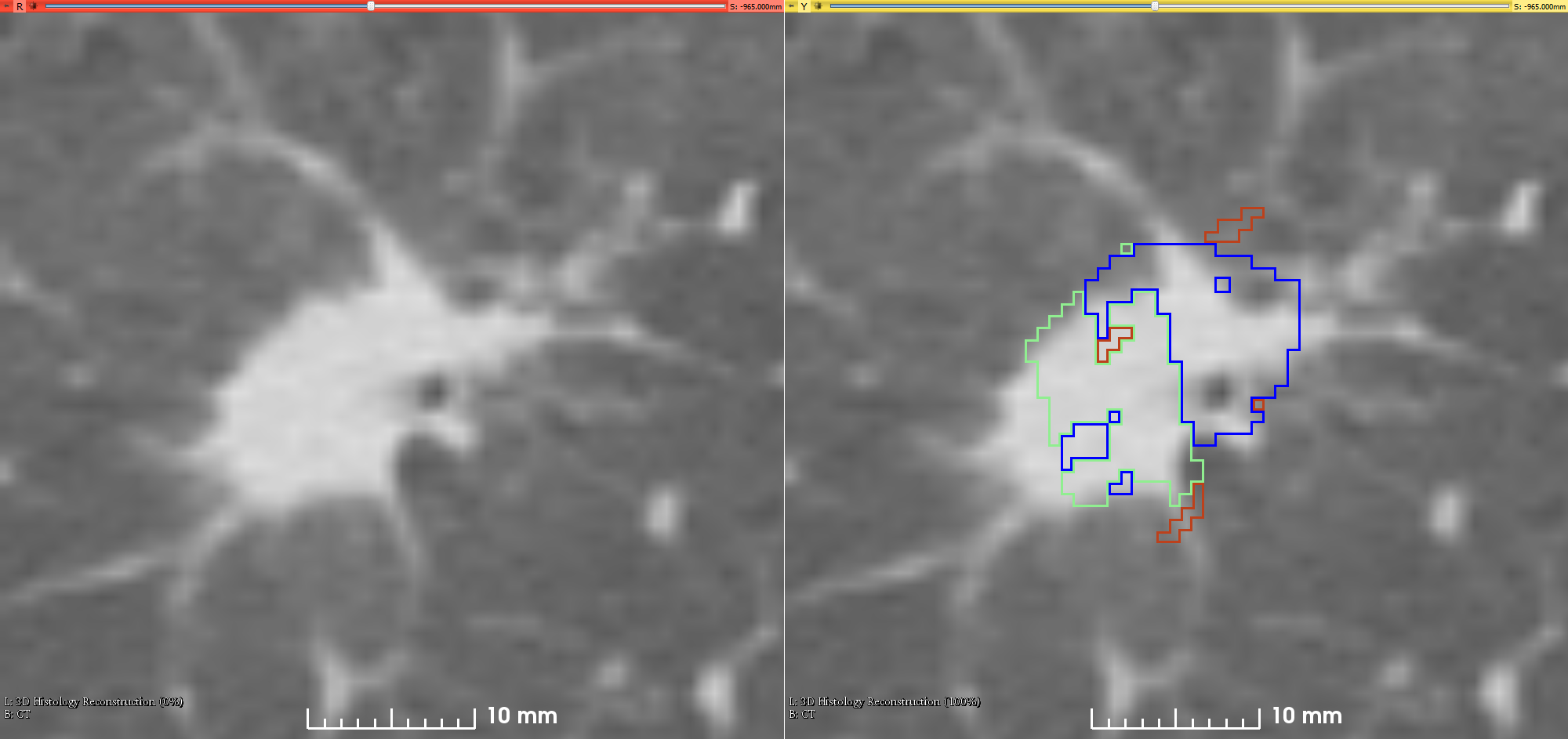
This is the first attempt of mapping the extent of Invasive Adenocarcinoma onto in vivo lung CT. The mappings constitute ground truth of disease and may be used to further investigate the imaging signatures of Invasive Adenocarcinoma in ground glass pulmonary nodules. Patient with small ground glass nodules with >2 histology slices per nodule were included. Patients with solid large nodules (>40mm), with <3...